Abstract
Background:
Whole-genome sequencing (WGS) costs are falling, yet, outside oncology, this information is seldom used in adult clinics. We piloted a rapid WGS (rWGS) workflow, focusing initially on estimating power for a feasibility study of introducing genome information into acute cardiovascular care.
Methods:
A prospective implementation study was conducted to test the feasibility and clinical utility of rWGS in acute cardiovascular care. rWGS was performed on 50 adult patients with acute cardiovascular events and cardiac arrest survivors, testing for primary and secondary disease-causing variants, cardiovascular-related pharmacogenomics, and carrier status for recessive diseases. The impact of returning rWGS results on short-term clinical care of participants was investigated. The utility of polygenic risk scores to stratify coronary artery disease was also assessed.
Results:
Pathogenic variants, typically secondary findings, were identified in 20% (95% CI, 11.7–34.3). About 60% (95% CI, 46.2–72.4) of participants were carriers for one or more recessive traits, most commonly in HFE and SERPINA1 genes. Although 64% (95% CI, 50.1–75.9) of participants carried at least one pharmacogenetic variant of cardiovascular relevance, these were actionable in only 14% (95% CI, 7–26.2). Coronary artery disease prevalence among participants at the 95th percentile of polygenic risk score was 88.2% (95% CI, 71.8–95.7).
Conclusions:
We demonstrated the feasibility of rWGS integration into the inpatient management of adults with acute cardiovascular events. Our pilot identified pathogenic variants in one out of 5 acute vascular patients. Integrating rWGS in clinical care will progressively increase actionability.
Introduction
Whole genomic sequencing (WGS) is anticipated to be a prerequisite for precision medicine but is rarely applied systematically in adult clinical settings outside of oncology.1 The potential for establishing mechanism and for patient stratification through genomic tools has yet to be fully realized, despite the rapidly falling cost for WGS.2 Adoption and implementation of WGS in routine clinical care are expected to lead to new discoveries through novel genotype-phenotype associations, to the identification of established or novel disease mechanisms, downstream diagnostics and therapeutics and a progressive evolution toward evidence-based precision medicine.3–6
Our knowledge of the genetic causes of cardiovascular disorders and the pharmacogenomics of many cardiovascular medications is constantly growing.7 Mendelian variants have been identified in around 50% of patients with hypertrophic cardiomyopathy or cardiac repolarization disorders, who may present with sudden cardiac death.8,9Although typically polygenic conditions, venous thromboembolism (VTE) and acute myocardial infarction (MI) may be initial presentations of hereditary thrombophilias10 or familial hypercholesterolemia,11 respectively. WGS can also reveal latent cancers that may present with unprovoked VTE or MI with available genotype-informed therapeutic options.12Currently, pharmacogenomics can guide treatment with anticoagulant, antiplatelet, statin, or other medications that are widely used in acute cardiovascular settings.7 As a single clinical test, WGS captures a depth of information, including Mendelian variants, pharmacogenomics, estimates of polygenic disease risk, and at sufficient read depth, somatic clonal disorders.13 Ultimately, it is proposed that WGS will underpin precision medicine, but without broad implementation, realizing this vision is unlikely to be straightforward.
There are several obstacles to the successful integration of genomics into current clinical workflow. Providers and patients often fail to recognize the hereditary nature of clinical traits. Genetic results are returned several weeks to months following the initial disease presentation and typically long after major clinical decisions have been made.14Genomic information must be available in a timely fashion for those making medical decisions if these data are ever to fully penetrate clinical workflows.15 Another barrier is the limited interpretation and interpretability of genetic data. Genotypes are often perceived as complex and nonactionable to practitioners and providers are unlikely to use information which they do not fully understand or are unable to act on themselves.14 As the drive to mainstream genomic testing continues, there is an impetus to develop supporting resources such as detailed, yet user-friendly, reports and to incorporate genomic data into guidelines, risk-assessment and clinical decision support.
Here, we evaluate a laboratory and clinical workflow for the return of rapid WGS (rWGS) results to providers and patients within the context of the index hospitalization. As proof of concept, we piloted a rWGS workflow in acute adult cardiovascular care, focusing initially on the feasibility of introducing genome information in this setting to enable timely clinical decision-making through real-time return of results. We outline an in-hospital service that combines streamlined consent, scalable family history collection, sample collection for multidimensional molecular phenotyping with the goal of timely return of actionable sequencing results to the patients and their physicians. The pilot establishes the necessary power for a definitive utility study to evaluate clinical and economic outcomes for rWGS across a broad range of adult diseases.
Methods
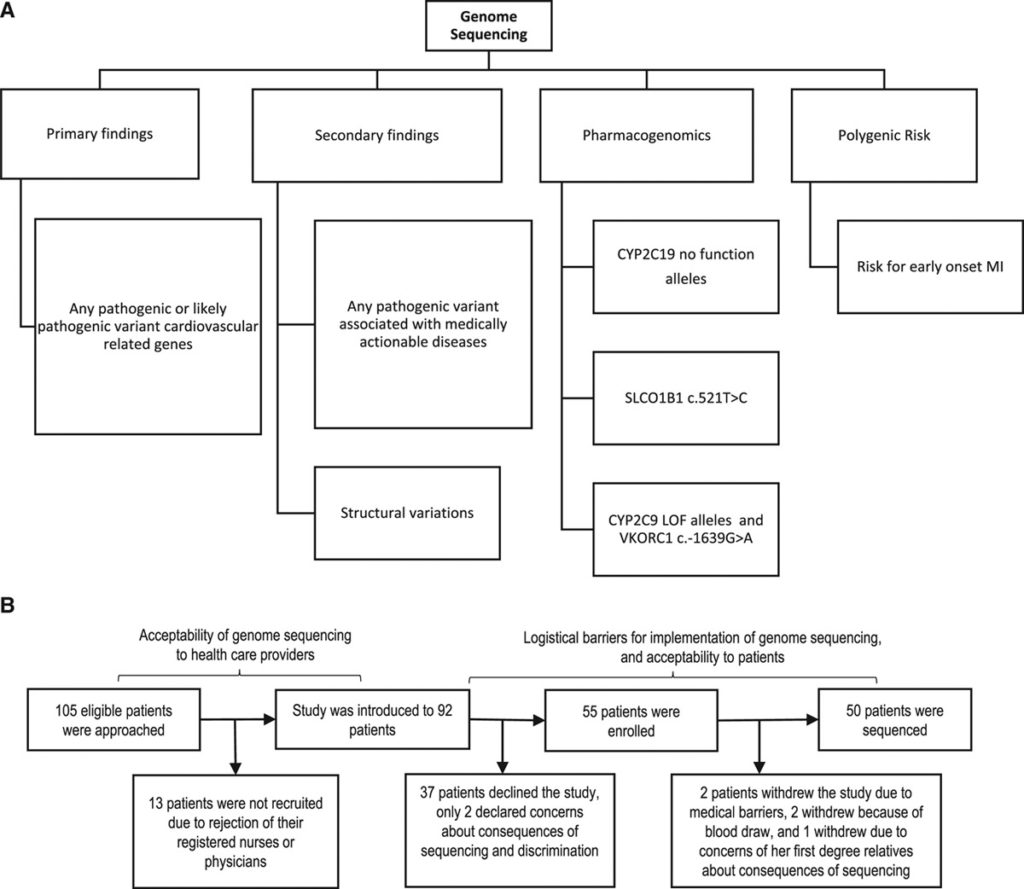
Institutional review board approval was obtained for this study. All patients and family members provided informed consent in accordance with the institutional review board protocol (no. 2014P002186). To protect privacy, each patient was assigned an anonymous identification number at admission. All participants agreed to receive primary and secondary findings and actionable pharmacogenomics information and signed the informed consent forms (Figure 1A). Anonymized patient-level data is publicly available according to the Transparency and Openness Promotion guidelines in the Data Supplement. However, the genomic data sets generated and analyzed during the current study are not publicly available due to data and privacy protection considerations but may be available on justified request to corresponding authors. A full description of the study protocol is found in the Methods and Tables I through V in the Data Supplement.
 Read the whole article at www.ahajournals.org
Read the whole article at www.ahajournals.org
